The Restriction Enzymes window is the place to control the selection and the filtering of restriction enzymes that will be used in the restriction analysis of DNA.
Opening the restriction enzyme window
Click on the main menu Tools > Restriction Enzymes or the contextual menu Sequence Analysis > Restriction Analysis > Restriction enzymes to open the Restriction Enzyme window (![]() ); the Properties window will open at the same time if not opened already.
); the Properties window will open at the same time if not opened already.
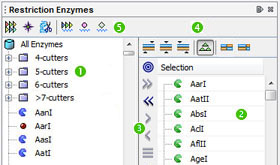
The following elements are visible in the window:
• Restriction enzyme panel (![]() ).
).
• Selection panel (![]() ).
).
• Selection toolbar (![]() ).
).
• Filtering toolbar (![]() ).
).
• Operation toolbar (![]() ).
).
The restriction enzyme panel
This panel presents to the user the list of Type II restriction enzymes available in GenBeans. Some special sets are predefined such as 'all enzymes' or '6-cutters'. Click on single enzymes or a set to select the corresponding restriction enzymes; use the Control key and the shift key at the same time to build complex selections. Click the Add button (![]() ) or the Replace All button (
) or the Replace All button (![]() ) in the selection toolbar to add or replace your new selection to the selection panel; these restriction enzymes are now marked with a red dot (
) in the selection toolbar to add or replace your new selection to the selection panel; these restriction enzymes are now marked with a red dot (![]() ) instead of the restriction enzyme icon (
) instead of the restriction enzyme icon (![]() ).
).
The selection panel
This panel exposes the set of selected restriction enzymes (![]() ) as a simple tree. When opened for the first time during a work session, the selection panel is populated with all prototypes with a cutting size superior or equal to 6.
) as a simple tree. When opened for the first time during a work session, the selection panel is populated with all prototypes with a cutting size superior or equal to 6.
The selection toolbar
This toolbar lets the user modify the set of selected restriction enzymes found in the selection panel.
• The Reset button (![]() ): Reset the selection panel with all restriction enzymes with a cutting size superior or equal to 6.
): Reset the selection panel with all restriction enzymes with a cutting size superior or equal to 6.
• The Replace All button (![]() ): Replace the current selection in the selection panel with all enzymes selected in the restriction panel.
): Replace the current selection in the selection panel with all enzymes selected in the restriction panel.
• The Remove All button (![]() ): Empty the selection panel.
): Empty the selection panel.
• The Add button (![]() ): Add the restriction enzymes selected in the restriction panel to the selection panel.
): Add the restriction enzymes selected in the restriction panel to the selection panel.
• The Remove button (![]() ): Unselect restriction enzymes from the selection panel.
): Unselect restriction enzymes from the selection panel.
• The Restrict button (![]() ): Restrict the selection panel to only those restriction enzymes that are also selected in the restriction panel.
): Restrict the selection panel to only those restriction enzymes that are also selected in the restriction panel.
• The Add Cutters button (![]() ): Replace the selection panel with all restriction enzymes that cut the current DNA sequence selection.
): Replace the selection panel with all restriction enzymes that cut the current DNA sequence selection.
• The Add Non-Cutters button (![]() ): Replace the selection panel with all restriction enzymes that do not cut the current DNA sequence selection.
): Replace the selection panel with all restriction enzymes that do not cut the current DNA sequence selection.
The filtering toolbar
This toolbar offers to the user varied filters to modulate the set of selected restriction enzymes.
• The 3' Protruding Ends Filter button (![]() ): When selected, only enzymes with 3'-protruding ends will be used.
): When selected, only enzymes with 3'-protruding ends will be used.
• The 5' Protruding Ends Filter button (![]() ): When selected, only enzymes with 5'-protruding ends will be used.
): When selected, only enzymes with 5'-protruding ends will be used.
• The Blunt Ends Filter button (![]() ): When selected, only enzymes with blunt ends will be used.
): When selected, only enzymes with blunt ends will be used.
• The Isoschizomers Filter button (![]() ): When selected, isoschizomers will be filtered out and only prototypes will be used (filter is active by default).
): When selected, isoschizomers will be filtered out and only prototypes will be used (filter is active by default).
• The Palidrome Filter button (![]() ): When selected, only enzymes with palindromic, symmetric recognition sites will be used.
): When selected, only enzymes with palindromic, symmetric recognition sites will be used.
• The Non-Palindorme Filter button (![]() ): When selected, only enzymes with non-palindromic, asymmetric recognition sites will be used.
): When selected, only enzymes with non-palindromic, asymmetric recognition sites will be used.
Restriction analyses will be performed on the current sequence selection in GenBeans using the current set of selected restriction enzymes.
• The Site Usage button (![]() ).
).
• The Restriction Map button (![]() ).
).
• The Digestion Report button (![]() ).
).
• The All Sites button (![]() ).
).
• The Unique Sites button (![]() ).
).
• The Absent Sites button (![]() ).
).