Protein Coding sequences or CDS are defined as sequences of nucleotides that correspond with the sequence of amino acids in proteins. Normally stop codons are included but GenBeans does not enforce this rule. Here is an example of representation:

Viewing CDS properties
To view the properties of a CSD feature, click anywhere in the CDS and lookup its properties in the Properties window:
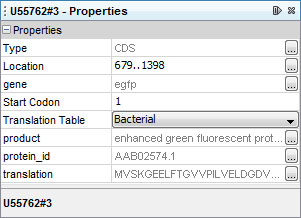
Some properties such as location, start codon and translation table can be edited directly in the Properties window.
Stop codon and fuzzy end representations
Special representation are used to show stop codons and fuzzy ends:
• Amber codon: ![]() for TAG (DNA) or UAG (RNA).
for TAG (DNA) or UAG (RNA).
• Ochre codon: ![]() for TAA (DNA) or UAA (RNA).
for TAA (DNA) or UAA (RNA).
• Opale codon: ![]() for TGA (DNA) or UGA (RNA).
for TGA (DNA) or UGA (RNA).
• Fuzzy end: ![]() for interrupted CDS or unknown end, see the feature table.
for interrupted CDS or unknown end, see the feature table.
Creating CDS
1. Select the part of the sequence who want to create a CDS from; possibly highlight first the translation to determine the proper limits of your protein sequence.
2. Click on New Feature button ( ![]() ) in the Viewer toolbar.
) in the Viewer toolbar.
3. Select the CDS subpanel in the Feature Panel:
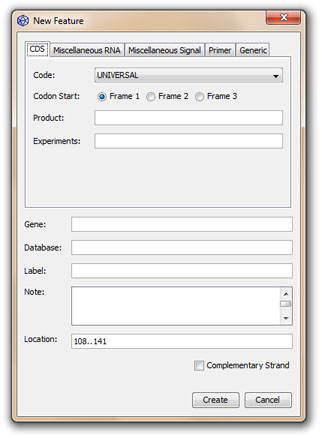
4. Fill up the different options if desired; for example choose the start codon (Frame 1, 2, or 3) or indicate the name of the protein product or add a note; possibly change the location (according to the feature table nomenclature) and finally click on the Create button to create the CDS feature.